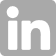An international team of scientists working with Heliconius butterflies at the Smithsonian Tropical Research Institute (STRI) in Panama was faced with a mystery: How do pairs of unrelated butterflies from Peru to Costa Rica evolve nearly the same wing-color patterns over and over again? The answer, published in Current Biology, forever changes the way evolution is understood.
An international team of scientists working with Heliconius butterflies at the Smithsonian Tropical Research Institute (STRI) in Panama was faced with a mystery: How do pairs of unrelated butterflies from Peru to Costa Rica evolve nearly the same wing-color patterns over and over again? The answer, published in Current Biology, forever changes the way evolution is understood.
“Our team is the first to report that although evolution of similar color patterns in Heliconius may be driven by similar forces—like predators avoiding a particular kind of butterfly—the pathway to that outcome is not predictable,” said Carolina Concha, lead author of the paper and a post-doctoral fellow at STRI. “This really surprised us because it reveals the importance of history and chance in shaping the genetic pathways leading to butterfly wing-pattern mimicry.”
Heliconius’ bright wing colors signal to bird predators that the butterflies are toxic. Flashy male wing patterns signal to females that they are choosing the right species to mate with. Somehow these two forces, predation and mating, lead to similar wing patterns in groups of butterflies isolated in the mountain valleys and foothills of the Andes. By knocking out a single gene called WntA in 12 different species and their variants, the molecular biologists on the team could tell whether the butterflies in a pair with the same wing patterns were using the same genetic pathways to color and pattern their wings. They were not.
Read more at Smithsonian Tropical Research Institute
Image: Carolina Concha. CREDIT: Smithsonian Tropical Research Institute/Carolina Concha