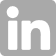In a world with a consistently growing population and a climate crisis, food shortage is a looming threat.
In a world with a consistently growing population and a climate crisis, food shortage is a looming threat. To alleviate this threat, crop scientists, botanists, genetic engineers, and others, have been exploring ways of boosting crop productivity and resilience. One way to control plant growth and physiology is to regulate the levels of "phytohormones" or plant hormones.
However, much remains to be known about the mechanisms that underlie this hormonal regulation in plants, limiting advancement in this direction. Now, in a study led by Nagoya University, Japan, a team of scientists has discovered, using rice plants as the study model, that a process called "allosteric regulation" is involved in maintaining the phytohormonal balance in plants. Their findings, published in Nature Communications, could hold the key to significantly advancing the research on plant growth and development, providing a potential solution for food security.
Plants survive by adapting their development and physiology to their surrounding environments by controlling the levels of enzymes driving the synthesis of two phytohormones, gibberellin and auxin. Enzymes are proteins that bind to one or more reactant chemicals and speed up a reaction process. The binding site is called the activation site. In 1961, it was discovered that in bacteria, enzyme activity is enhanced or inhibited via allosteric regulation, which essentially is the binding of a molecule called the "effector" at a site other than the active site of the enzyme. In allosteric regulation, the structure of the enzyme changes to either support or hinder the reaction that the enzyme enables.
Read more at Nagoya University
Image: Dimerization of OsGA2ox and OsDAO enhances its enzymatic activity. Under low substrate concentration (GA4 or IAA) (left), GA2ox or DAO functions as a monomer with low enzyme activity. At high substrate concentration (right), the interface GA4 or IAA causes the formation of multimers by bridging two enzyme molecules, resulting in hyper-activation. GA4 or IAA is retained in a stable interface position, allowing two subunits to enter the active site for the next reaction without a high energy barrier. Lys or Arg is the most important amino acid for retaining GA4 or IAA and for entering the active site. Furthermore, MD simulation of OsGA2ox3 revealed the presence of a gate, allowing substrate to enter the active site and for product to exit. This gate had a hinge site composed of three amino acids, W106, C186, and V196, and was also stabilized by the interaction between R97 in subunit A and F100 in subunit D. GA4-dependent dimerization enhanced its enzymatic activity. These mechanisms are conserved in all rice GA2oxs. (Credit: S.Takehara et al. 2020)