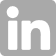While sifting through the bacterial genome of salmonella, Cornell food scientists discovered mcr-9, a new, stealthy jumping gene so diabolical and robust that it resists one of the world’s few last-resort antibiotics.
While sifting through the bacterial genome of salmonella, Cornell food scientists discovered mcr-9, a new, stealthy jumping gene so diabolical and robust that it resists one of the world’s few last-resort antibiotics.
Doctors deploy the antibiotic colistin when all other infection-fighting options are exhausted. But resistance to colistin has emerged around the globe, threatening its efficacy.
“This last-resort antibiotic has been designated a highest-priority antibiotic by the United Nations’ World Health Organization, and the mcr-9 gene causes bacteria to resist it,” said Martin Wiedmann, Cornell’s Gellert Family Professor in Food Safety and senior author on the study published May 7 in the journal Mbio. “In treatments, if colistin does not work, it literally could mean death for patients. If colistin resistance spreads, a lot of people will die.”
Co-lead author Laura Carroll, a computational biologist and Cornell doctoral candidate, found mcr-9 in the genome of a strain of foodborne pathogen salmonella.
Read more at Cornell University
Image: The mcr-9 gene's protein structure graphic depicts the differing protein chains (solid color ribbons) and the protein surface (mesh) surrounding it. CREDIT: Ahmed Gaballa/Cornell University